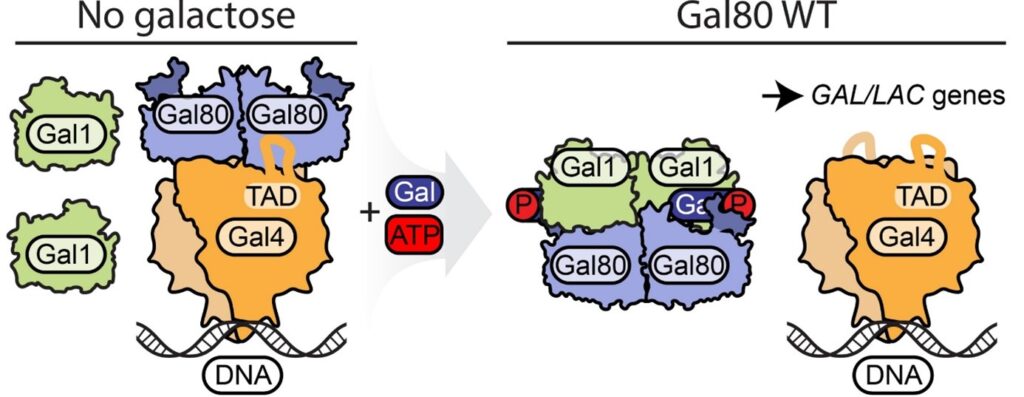
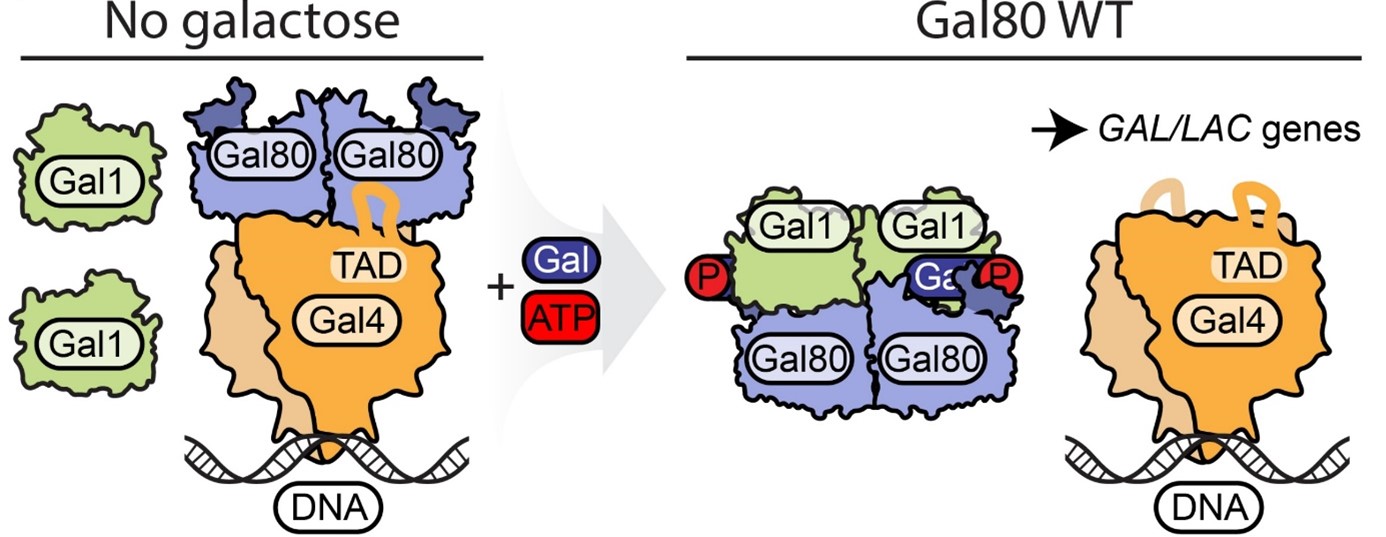
To grow efficiently, yeast often need to swich from glucose to galactose and backwords. This is why they developed a simple, yet very efficient mechanism composed of three proteins that turns on when galactose is around and enables the switch to an alternative carbon source. In Saccharomyces cerevisiae GAL operon is composed of Gal4, Gal80 and Gal3, which in Kluyveromyces lactis is named Gal1. To cut a long story short, Gal1 is a galactose kinase which senses galactose concentration and strives to activate Gal4. Gal4 is a transcription factor responsible for expression of genes related to galactose metabolism. To prevent spontaneous activation, there is a Gal80 protein, which acts as a suppressor of this mechanism. Until now Gal80 was supposed to act in the same way in S. cerevisiae and K. lactis.
Thanks to the joint efforts of international teams from Martin Luther University (Halle Wittenberg, Germany) and Max Planck Research Group (Kraków, Poland) now we know, that there are significant differences between S. cerevisiae and K. lactis Gal80 proteins. Firstly, unlike yeast Gal80, K. lactis Gal80 on its N-terminus contains a nuclear localization signal, which allows to piggyback Gal1 to the nucleus. Secondly, the disordered N-terminus of Gal80 is involved in direct interaction with Gal1, as shown in biochemical and computational docking experiments. Altogether, our findings published in Life Science Alliance shed light on the induction of GAL operon, a textbook example of regulation of gene expression in Eukaryota.
You can read the publication here: https://www.life-science-alliance.org/content/3/12/e202000665
DOI 10.26508/lsa.202000665