Dr. Ting Yu-Lin from our Team has been awarded with a Sonata grant from National Science Center for the realization of her project “Molecular and structural insight into the human pseudouridine synthase 3”. Congratulations! You can read more about her project down below.
How do pseudouridine synthases select their substrates?
RNA consists of four basic elements and diverse chemical modifications expands its structural capacity greatly. Pseudouridine is one of the most abundant modification that can be found in all kinds of RNAs, such as tRNAs and rRNAs. The presence of pseudouridine strengthens RNA-RNA or RNA-protein interaction, that involves in many biological regulations. Recently, mapping pseudouridine in a transcriptome-wide analysis has even revealed that pseudouridines are also present in mRNAs, which adds a new layer of regulation.
The pseudouridylation is catalysed by pseudouridine synthases (Pus) that one class of enzyme recognize their substrates by the guide RNA whereas the other class is stand-alone enzymes. The later class contains several Pus enzymes, including Pus1-10. Several reports have demonstrated some Pus crystal structures from human or other model organisms. They all share the similar protein folding and a conserved catalytic centre despite the low sequence similarity. Moreover, each Pus enzyme has different sets of pseudouridylation targets. For instance, Pus4 and Pus7, recognize substrate by consensus sequences while others, such as Pus1, target the substrates by their secondary structures. Pus3 belongs to the same subfamily as Pus1 yet they catalyse pseudouridylation on different sites of targets. It raises the question that how these enzymes determine their substrate targets. In addition, mutations in Pus enzymes have been linked to onset of diseases however the underlying mechanism still needs to be investigated.
We have proposed to obtain Pus3’s structure using either a crystallography approach or a cryoEM strategy. Furthermore, we will characterize Pus3’s biophysical and biochemical activities toward different RNA substrates and compare them with other Pus enzymes. We believe gathering all essential observations could expand our knowledge on the molecular details of the Pus enzyme and the basis may provide the pathogenic link to disease formation.
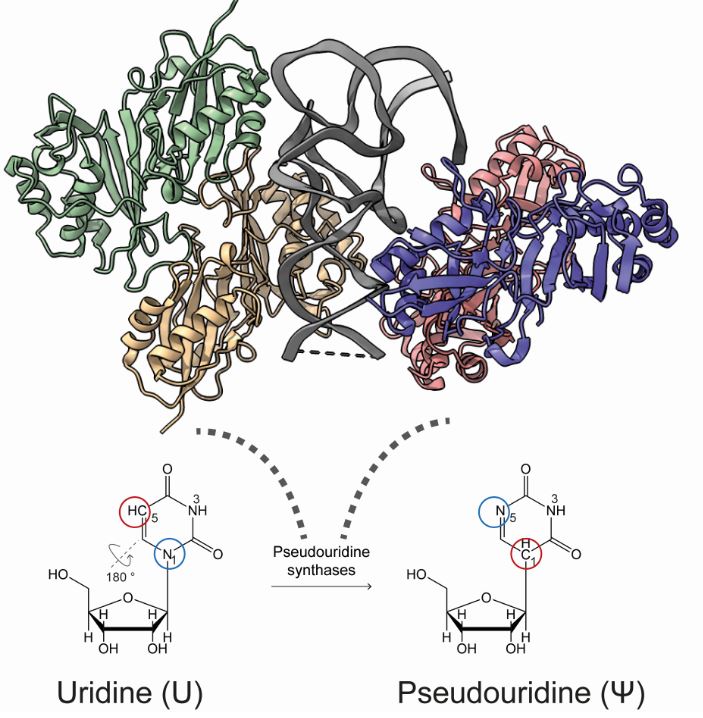
A scheme of conversion of uridine to pseudouridine by a pseudouridine synthase (Pus) (modified from PDB: 2NQP). The flipped sites are colored coded. The tRNA in the co-crystal is color in grey while the individual chain of E. coli TruA is colored differently.