Pseudouridylation is a crucial nucleic acid modification that affects the structure and function of almost all cellular RNA molecules. Recently, pseudouridines gained particular attention, due to their utility for mRNA SARS-CoV2 vaccines. While the modified version of uridine was discovered more than 60 years ago, around the same time as the structure of DNA, we still know very little about the enzymes that can introduce this modification into our RNAs. In their latest work, scientists from the Jagiellonian University and the University of Bern presented a detailed molecular description of the vital human enzyme called pseudouridine synthase 3 (PUS3). The study, recently published in Molecular Cell, shows how the human PUS3 recognizes, binds, and modifies specific sites in transfer RNAs (tRNAs).
The scientists from the Max Planck Research Group at the Malopolska Centre of Biotechnology (MCB) of the Jagiellonian University (JU) in Krakow determined the first cryo-electron microscopy (cryo-EM) structures of the long-neglected human enzyme PUS3. While in the past work, the group linked specific patient-derived mutations in PUS3 with rare human diseases, the current insights allow us to understand the mechanisms behind these links.
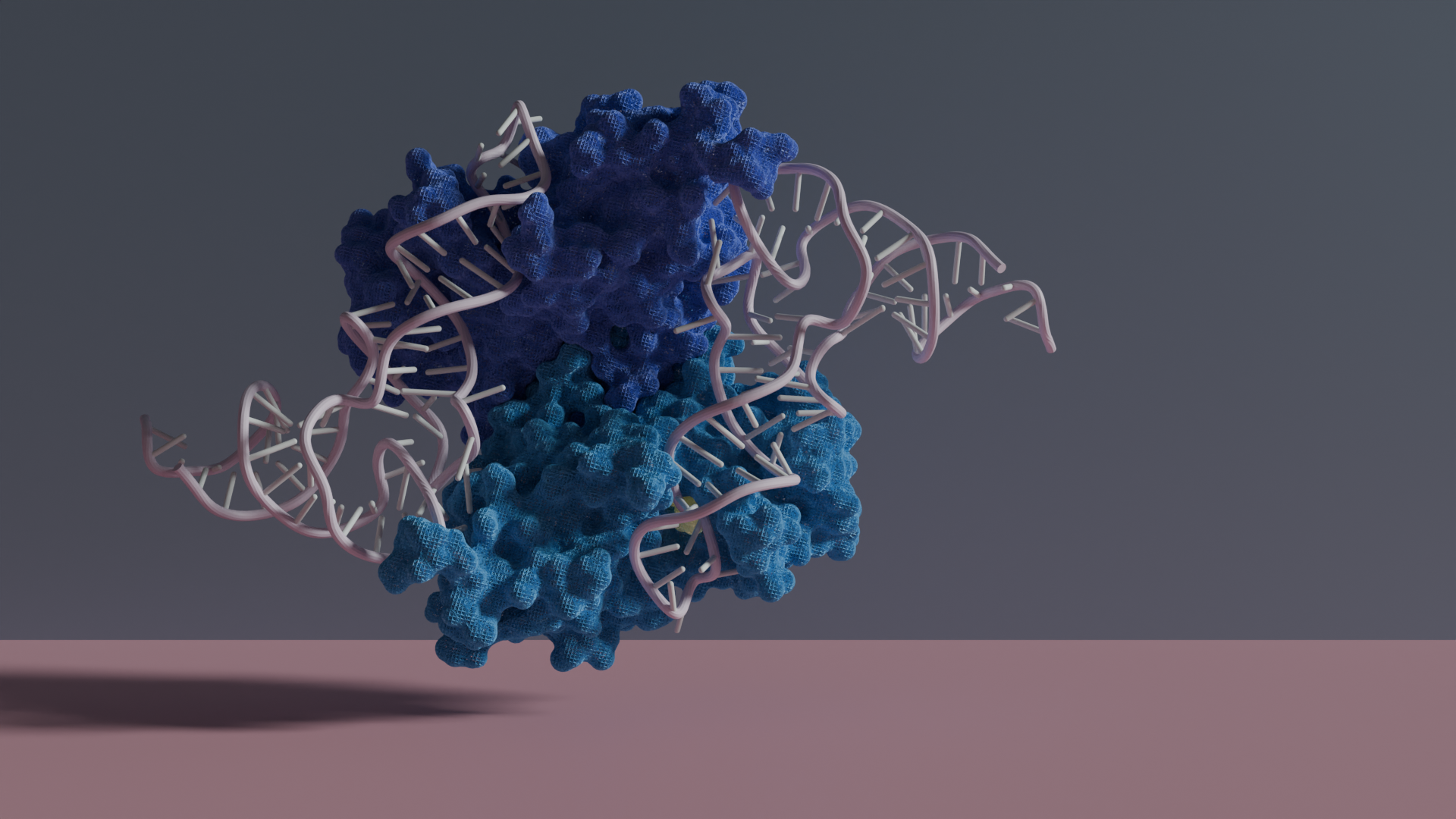
The current study reveals not just the structure of the resting human PUS3, which is ‘waiting’ for its target substrate, but also in complex with different tRNA molecules. This gallery of structures shows that two identical PUS3 enzyme molecules form a so-called homodimer, which is crucial for both the structural integrity of the protein and its capability to bind tRNAs. Only in this homodimer configuration, the correct uridine is positioned near the active site of one PUS3 monomer, while another region of the tRNA contacts the second monomer. Therefore, the cryo-EM structures illustrate how PUS3 accurately places the target uridine in the catalytic site for the subsequent modification reactions – explaining its specificity and mechanism at the atomic level. To better understand the prevalence of pseudouridylation sites in the cell, the Leidel group in Switzerland used engineered human cell lines and they confirmed that PUS1 and PUS3 target different sets of cellular RNAs. While PUS1 targets a wide range of RNAs, including tRNAs and messenger RNAs (mRNAs), the PUS3 specifically modifies only tRNAs. Last but not least, the authors established a reporting system to express clinically relevant PUS3 variants using human cell lines, enabling them to monitor and confirm the impacts of different clinical mutations on the activity of the enzyme. These cell culture experiments were carried out in collaboration with the neighbouring Faculty of Biochemistry, Biophysics, and Biotechnology (WBBB) at the JU.
“Pseudouridine synthases were discovered decades ago, and we are excited that the advent of new techniques helps us expand our knowledge of the PUS3 homodimer complex and its enzymatic activity as well as how it achieves substrate selection” comments the shared first author of the study, Dr. Ting-Yu Lin. “We looked at PUS3 from several perspectives, which allows us to draw a conclusive overall picture of this clinically highly relevant human enzyme” explains Dr hab. Sebastian Glatt, the shared corresponding and last author of the work.
All structural biology experiments were conducted with the strong support of the Structural Biology Core Facility at MCB. All cryo-EM data were collected on the Titan Krios G3i, a high-end cryo-electron microscope, located at SOLARIS National Synchrotron Radiation Centre. The work at MCB was supported by an ERC Consolidator grant that is currently implemented in the group of Dr. Glatt.