All living organisms use information coded in their genomes (DNA) to produce proteins, including all active enzymes. These complex molecular machines are responsible for carrying out most of the fundamental cellular functions, like cell division, production of energy, sensing of the environment and reaction to external stimuli. During a process called “transcription” regions that are the blueprints for specific proteins (genes) are activated and give rise to messenger ribonucleic acid (mRNA) molecules. These mRNA molecules contain a copy of the sequence information stored in the DNA and in higher organisms are transported out from the cell nucleus into the cytoplasm. Next, ribosomes and transfer RNA (tRNA) molecules convert the encoded information from mRNAs into correctly assembled chains of linked amino acids in a process called “translation”. These chains fold into specific three-dimensional structures and then can get called proteins. Of note, the specific function and enzymatic activity of a protein depends on their intrinsic properties and structural features. All steps of transcription and translation are tightly controlled to assure the correct production of the right enzymes at the right time in the right cellular context. In addition, the folding of a poly peptide chain is a highly dynamic and complicated multi-step process that needs tight regulation to assure correct assembly and functionality of the produced enzymes. Mutated or incorrectly folded proteins, inappropriate expression levels as well as the production of a protein in the wrong cellular context can lead to cellular dysfunction and in the worst case promote the development of severe diseases, like cancer or epilepsy.
In our research group, we aim to understand fundamental cellular mechanisms that are highly conserved from yeast to humans and affect protein synthesis at the level of translation. In detail, cells use a multi-protein machines to attach small chemical modifications to mRNAs and tRNAs, which guarantee that proteins are produced with highest precision and at the appropriate speed to guide correct three-dimensional folding. Our work from previous years has mainly focused on understanding protein complexes that conduct specific RNA modifications higher organisms.
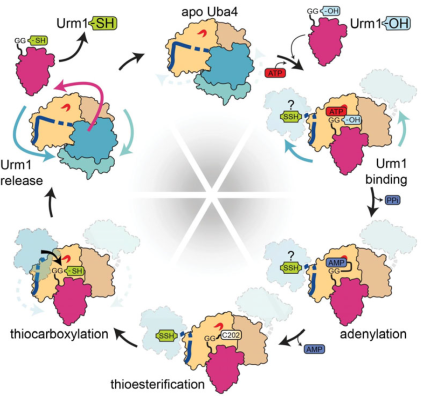
In the proposed project, we will investigate a very specific cellular pathway that is needed to incorporate sulfur atoms into tRNAs. Over the last years, we have found out that in yeast sulfur is exclusively relayed via a cascade of several proteins to the C-terminal end of one single unique protein, called Urm1. Subsequently, Urm1 is able to transfer this activated sulfur atom not only to tRNAs, but also to other target proteins. Strikingly, humans also possess a second protein, called MOCS2A, that also carries a sulfur atom at its C terminus. This protein does not seem to be capable of transferring sulfur to tRNAs or proteins, but it is needed during the biosynthesis of chemical compound called molybdenum cofactor (Moco). This co-factor requires the incorporation of several sulfur atoms, before it is itself incorporated into so called molybdoenzymes. Deficiencies that affect Moco synthesis triggers various human disease, including neurological abnormalities and intellectual disability.
Strikingly, MOCS2A as well as URM1 are still activated by the same enzyme in humans, namely MOCS3. In the proposed project we will shed light on the specific molecular similarities as well as differences between the two sulfur carrier proteins and their common activator.
We will purify the proteins and analyze their molecular characteristics and activities in the test tube. We will use structural biology methods to take structural snapshots of the proteins during their reactions to understand their function. Last but not least, we seek to identify the networks and natural targets of both proteins in human cell lines that will also serve as simplified model systems to understand the contribution of these proteins to human diseases
The expected results from the proposed research plan will provide deep molecular insights into yet poorly characterized regulatory networks of protein homeostasis and cellular sulfur metabolism, which have strong clinical implications in severe human diseases. Therefore, the work is essential to define the proteins of interest as potentially interesting diagnostic markers and intervention points for future targeted therapies.