Every living organism is composed of cells – “miniature” factories that ensure balance between health and disease in a process called homeostasis. Among the guardians of cell homeostasis are protein enzymes that perform versatile functions. One of them is guiding RNA molecules (messenger RNA – mRNA and transfer RNA – tRNA) to properly translate encoded genetic information into correctly folded polypeptide chains. There are over 170 unique modifications found in RNA molecules that modulate their function and act as additional level of translational regulation. The exact role of different RNA modifications is not fully understood and arouses curiosity of many scientists worldwide.
It is well known that RNA consists of four nucleobases (namely C, U, A, G), that may be decorated through adding chemical modifications by protein enzymes to modify their function in various cell types. Dihydrouridine is a small chemical change of U base, frequently found in RNA loop regions, which has been shown to locally loosen bonds between RNA bases. The U to D conversion requires RNA modifying enzymes, namely dihydrouridine synthases (DUS). There are four human DUS enzymes, each of them alters tRNA in specific positions. Any disfunction of their action leads to severe consequences. Higher levels of DUS proteins have been linked with poor prognosis of lung cancer patients or the onset of Alzheimer’s disease.
In this project we aim to characterize human DUS enzymes bound to tRNAs to understand how they recognize specific tRNAs molecules and how to moderate their activity.
The proposed project is designed to address and directly answer the following research tasks:
The proposed study will follow a well-designed research plan to obtain a comprehensive molecular picture of the interactions between human DUS enzymes and their substrates. We aim to translate this knowledge by identifying pan- and/or specific lead compounds that can modulate activity of human DUS enzymes.
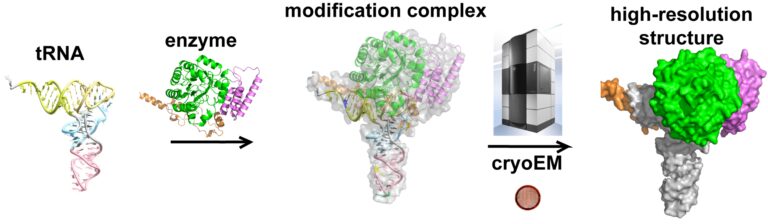
Despite major scientific efforts, many of the molecular consequences of RNA modifications remain poorly described. To understand how DUS enzymes achieve specificity for various tRNA targets, detailed characterization of their three-dimensional architecture and their relationships, so called ‘interactions’ with tRNA is required.
We have chosen a state-of-the-art technique of structural biology – cryo-electron microscopy (cryo-EM) which will allow us to ‘observe’ high resolution details of DUS-tRNA molecules for the first time. Obtained results will be used to build structural maps to visualize their atomic interactions and describe catalytic reaction mechanism. Next, we will use the structural information to measure enzymes activity, binding to tRNA and stability of modified tRNA in thorough biochemical and biophysical analyses.
These results will help us pinpoint the details of how a given DUS can select its specific tRNA partner and perform the dihydrouridine synthesis. Moreover, we will use two crystal structures of DUS enzymes to seek small molecules that may inhibit or modulate DUS activity. Finding the very first DUS inhibitors will broad our knowledge about DUS function and pave the way for development of novel therapeutic drugs for incurable diseases like cancer.
Broadening our understanding of how DUS enzymes select and convert key uridines on its target tRNAs to alter their role in protein production, has great scientific value and potential medical application. Out research will facilitate the development of targeted therapeutic strategies for lung cancer patients or individuals with onset of neurodevelopmental diseases.