Protein synthesis is heavily affected by the presence of various modifications in tRNAs. Wobble uridines in the anticodon stem loop are modified by a large cellular machine called Elongator. Genomic mutations in this macromolecular complex lead to the onset of severe human diseases. The latest work of scientists from the Malopolska Centre of Biotechnology (MCB) of the Jagiellonian University describes the first high-resolution cryo-electron microscopy (cryo-EM) structures of human Elongator and its mechanism of action. The study was published recently in Nature Communications.
Scientists from the Max Planck Research Group at the MCB of the Jagiellonian University in Krakow fulfilled their longtime goal of understanding how human Elongator works. The research of dr hab. Sebastian Glatt’s team was performed in collaboration with the scientists from Kassel University and Berlin Technical University.
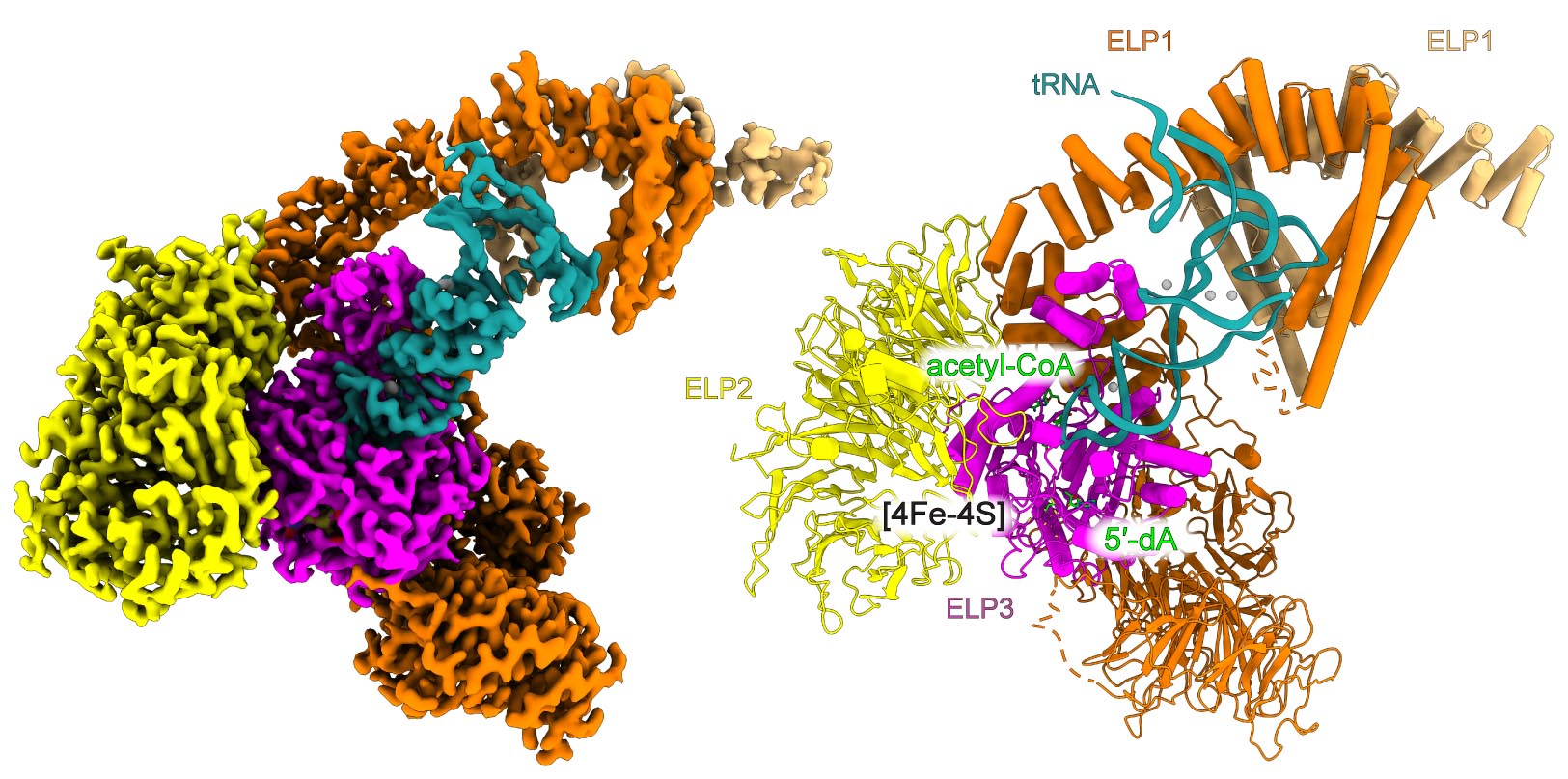
The multi-subunit Elongator complex is composed of two distinct subcomplexes, namely Elp123 and Elp456. The carboxymethyl group (cm5) introduced by Elongator serves as the basis for other enzymes for the synthesis of subsequent modifications. tRNAs with fully modified wobble uridines bind to translating ribosomes in an optimal manner and thus facilitate effective co-translational folding of emerging proteins. Structural biology and in vitro experiments were performed at MCB, with the assistance of the Structural Biology and Proteomics and Mass Spectrometry Core Facilities at MCB. Cryo-EM data was collected on the Titan Krios G3i, a high-end cryo-electron microscope located at SOLARIS National Synchrotron Radiation Centre. The team from Kassel conducted in vivo analyses, while the team from Berlin performed crosslinking mass spectrometry experiments.
The MCB team resolved the structure of the human Elongator complex at the highest resolution among all Elongator structures (2.9 Å), which is published so far. The structure shows human ELP123, in complex with tRNA and acetyl-CoA molecule. It precisely depicts the organization of the active site together with the tRNA anticodon stem loop, and modification target, namely the wobble uridine. The structure allowed to identify the unforeseen role of another universally conserved uridine adjacent to the wobble position which is important for the activity of ELP123. This collaborative work presents structural snapshots of the complex during intermediate stages of modification reaction. The authors also identified a series of conserved amino acid residues located in the active site of the catalytic subunit, which are necessary for the chemical reaction. All these findings were verified and supported by in vivo and in vitro experiments.
“It is exciting that, even with the extensive exploration of Elongator in the past, the advent of new techniques helps us challenge our knowledge of the complex and its enzymatic activity.” comment the two shared first authors of the study, Dr. Nour-el-hana Abbassi and Dr. Marcin Jaciuk. Dr hab. Sebastian Glatt, the leader of the team in Krakow, states that “it was always our goal to understand how this clinically important protein complex works in humans. Our high-resolution structure of human Elp123, allows us to precisely and directly pinpoint positions of the mutations that lead to Elongator associated disorders and try to understand their impact on the Elongator activity.”
The work was supported by the European Research Council under the European Union’s Horizon 2020 and innovation programme (101001394) and the Foundation for Polish Science (TEAM TECH CORE FACILITY/2017-4/6)